Proteins and Enzymes
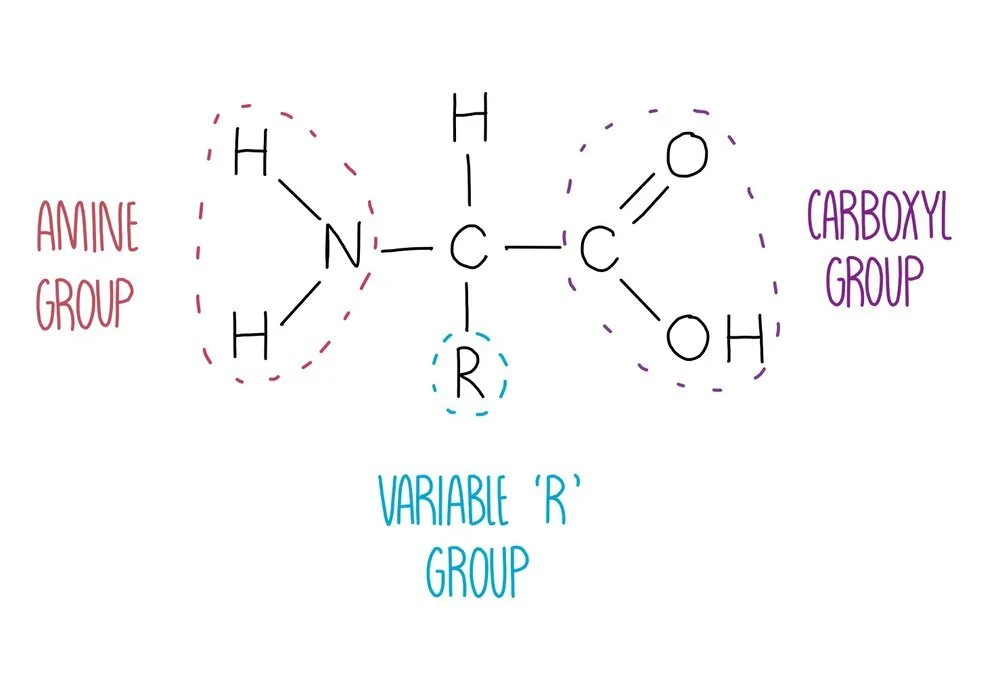
Amino acid structure
Proteins are polymers of amino acids joined together by peptide bonds. There are 20 different amino acids that our body needs to stay healthy, all of which have the same general structure. They consist of a central carbon atom attached to four different groups: an amine group, a hydrogen atom, a carboxyl group and an 'R’ group which is different in each amino acid. The identity of the R group will influence how the amino acid interacts with other amino acids, therefore influencing protein folding. For example, lysine has a charged R group and is able to form ionic bonds with negatively charged amino acids.
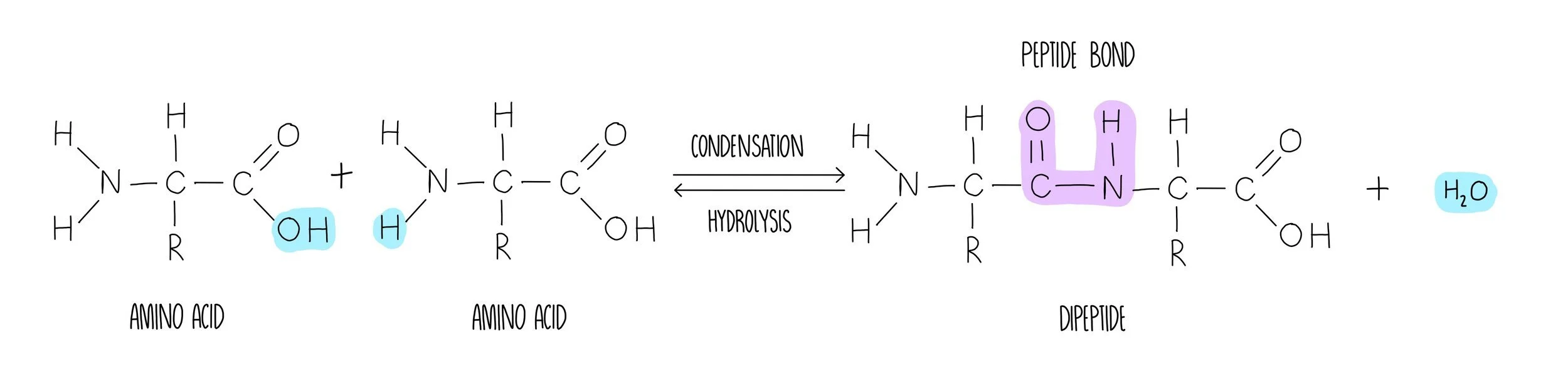
Peptide bond formation
Peptide bond formation involves the removal of water in a condensation reaction. The water is formed by removing a hydrogen atom from the amine group and a hydroxyl (-OH) group from the carboxylic acid. What’s left behind is a peptide bond (-CONH) which can be broken with the addition of water in a hydrolysis reaction.
Protein structure
Once a long polypeptide chain is synthesised from the formation of numerous peptide bonds between adjacent amino acids, the polypeptide undergoes multiple stages of folding. These stages of protein origami are defined as the primary, secondary, tertiary and quaternary structures which describe a protein from its most unravelled to its final compact structure.
Primary structure: peptide bonds have formed between amino acids to form a long, straight chain (polypeptide).
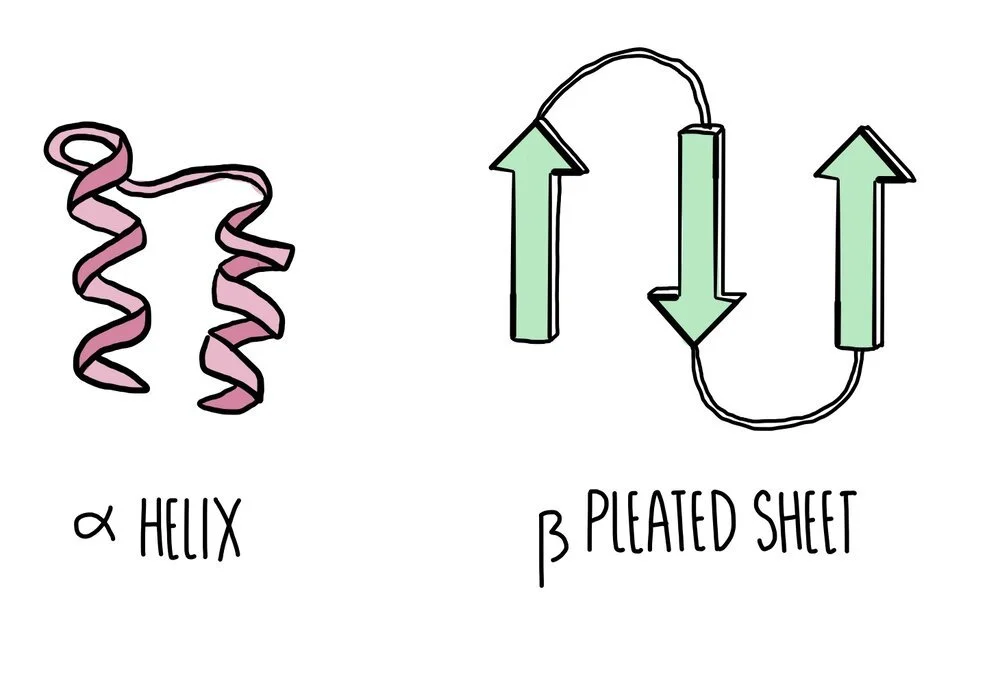
Secondary structure: hydrogen bonds form between nearby amino acids (from the amine group on one amino acid to the carboxyl group of another) to form either an alpha helix or a beta pleated sheet. Proteins which form neither of these two structures will form a random coil.
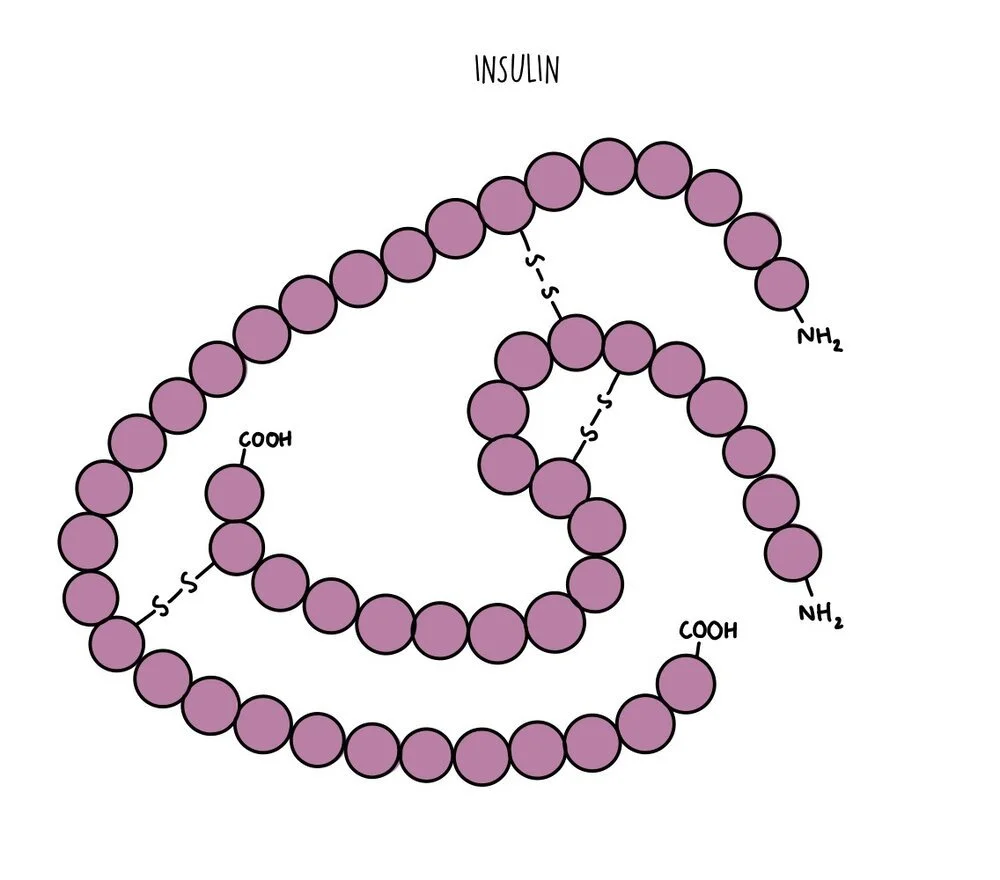
Tertiary structure: more bonds form between the different R groups to give the protein a 3D structure. R-group interactions involve hydrogen bonds, disulfide bonds, ionic bonds and polar interactions. If proteins are made of a single polypeptide chain, this is their final overall structure.
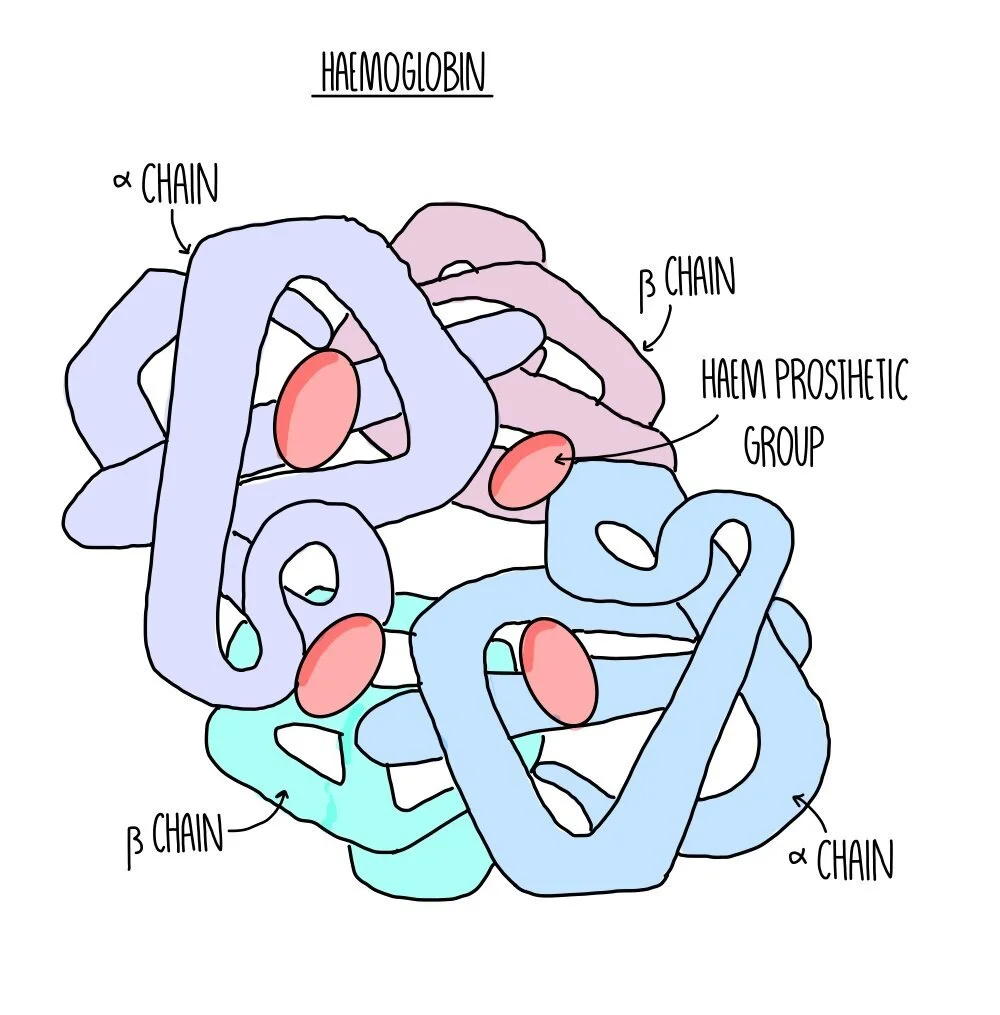
Quarternary structure: this is the structure formed from the interaction of multiple polypeptide chains held together by bonds. Haemoglobin is an example of a protein with quaternary structure. It consists of four polypeptide chains (two alpha chains and two beta chains) bonded together. Each chain surrounds an iron-containing haem group. The haem group is referred to as a prosthetic group - non-protein components which are required for protein function.
Globular vs fibrous proteins
All proteins can be organised into one of two broad categories: globular proteins and fibrous proteins. Globular proteins are spherical and arranged with their hydrophobic amino acids tucked inside and the hydrophilic amino acids exposed on the outside. This means globular proteins are soluble and can be transported easily from one part of the cell to another. They perform functional roles and include things like enzymes (such as amylase), hormones (such as insulin) or proteins like haemoglobin. Globular proteins unravel and denature when the temperature or pH deviates from optimum levels. Globular proteins with prosthetic groups attached (such as haemoglobin) are referred to as conjugated proteins.
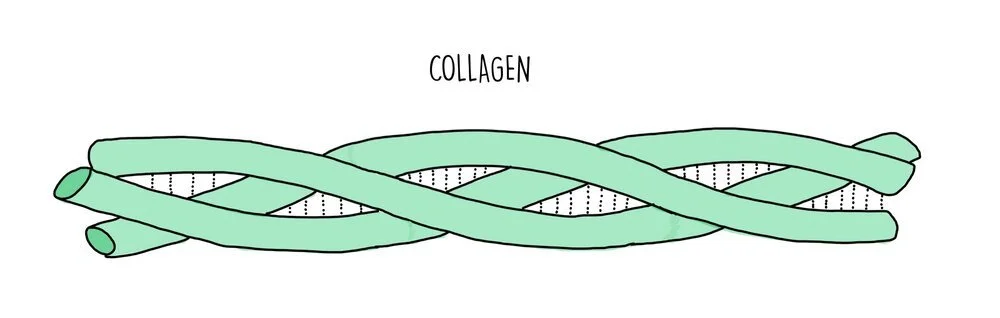
Fibrous proteins are long and thin and their primary structure consists of a repetitive sequence of amino acids. They perform structural roles so they are strong and insoluble. Examples of fibrous proteins include collagen, keratin and elastin. Collagen consists of three polypeptide chains wrapped tightly around each other to form a stable quaternary structure held together by numerous hydrogen bonds. Collagen is found in connective tissue, such as skin, muscle and bone. Fibrous proteins tend to be less sensitive than globular proteins to changes in temperature and pH.
Enzyme function and structure
Enzymes are biological catalysts - they speed up the rate of chemical reactions happening inside our body. They work by reducing the activation energy of a reaction. Activation energy is defined as the minimum amount of energy needed for a reaction to happen. If less energy is needed, then reactions can take place as lower temperatures than would be needed without an enzyme. Without the enzymes in our bodies, the reactions that happen inside of us would not be possible at normal body temperature. Remember that enzymes are unchanged at the end of a reaction which means they can be reused.
Enzymes can be classed as either intracellular if they catalyse reactions inside cells e.g. RNA polymerase, or extracellular if they catalyse reactions outside of cells e.g. amylase. All enzymes are globular proteins and have regions called active sites. The active site of an enzyme has a specific shape and allows the substrate to bind. Other enzymes may have regulatory regions where an inhibitor can bind, which we refer to as the allosteric site.
Mechanisms of enzyme action
Scientists have two ideas to explain the way in which enzymes work: the ‘lock-and-key’ model and the ‘induced-fit’ model. They are models because they are our best-accepted theories based on the evidence we have available.
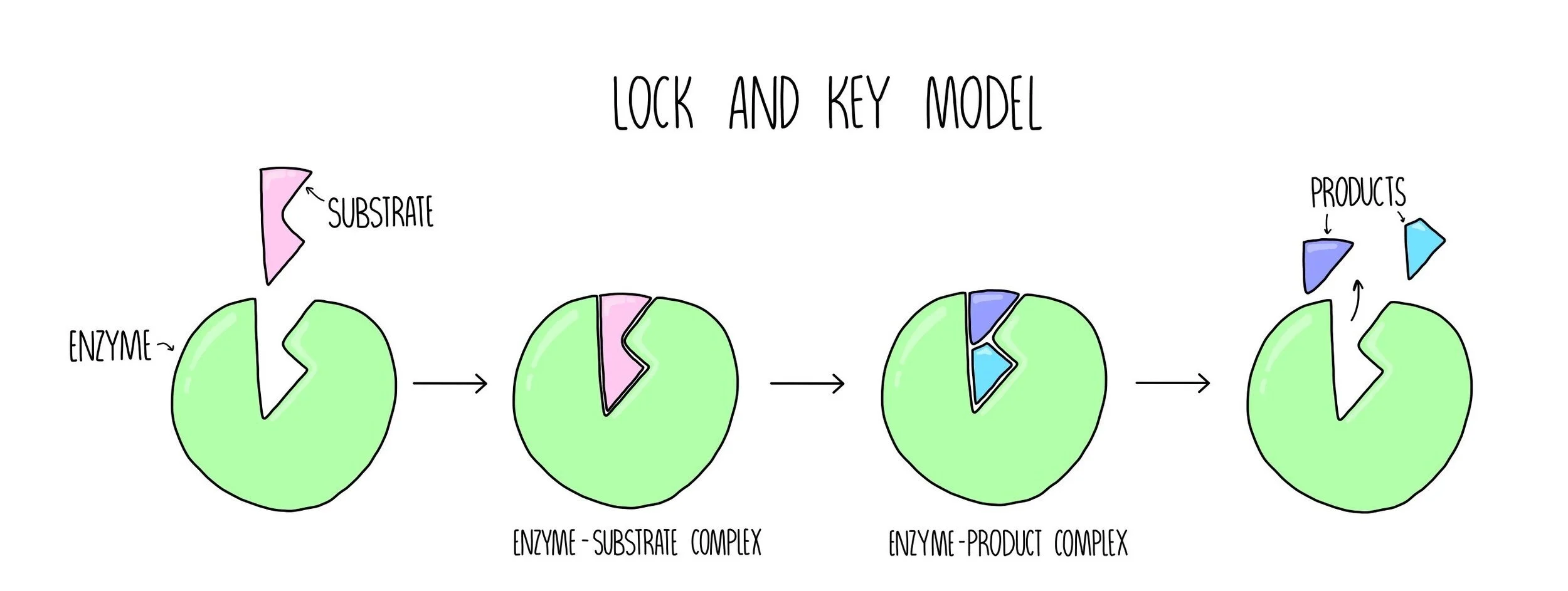
Lock and Key model
The lock and key model is the simpler of the two theories of enzyme action. This model suggests that the substrate fits into the enzyme’s active site in the same way in which a key fits into a lock. The shape of the substrate and the active site are perfectly complementary to each other. Catalysis happens in the following stages:
The substrate binds to the enzyme’s active site, forming an enzyme-substrate complex (ES complex).
The enzyme converts the substrate into product, forming an enzyme-product complex (EP complex).
The product is released from the enzyme’s active site.
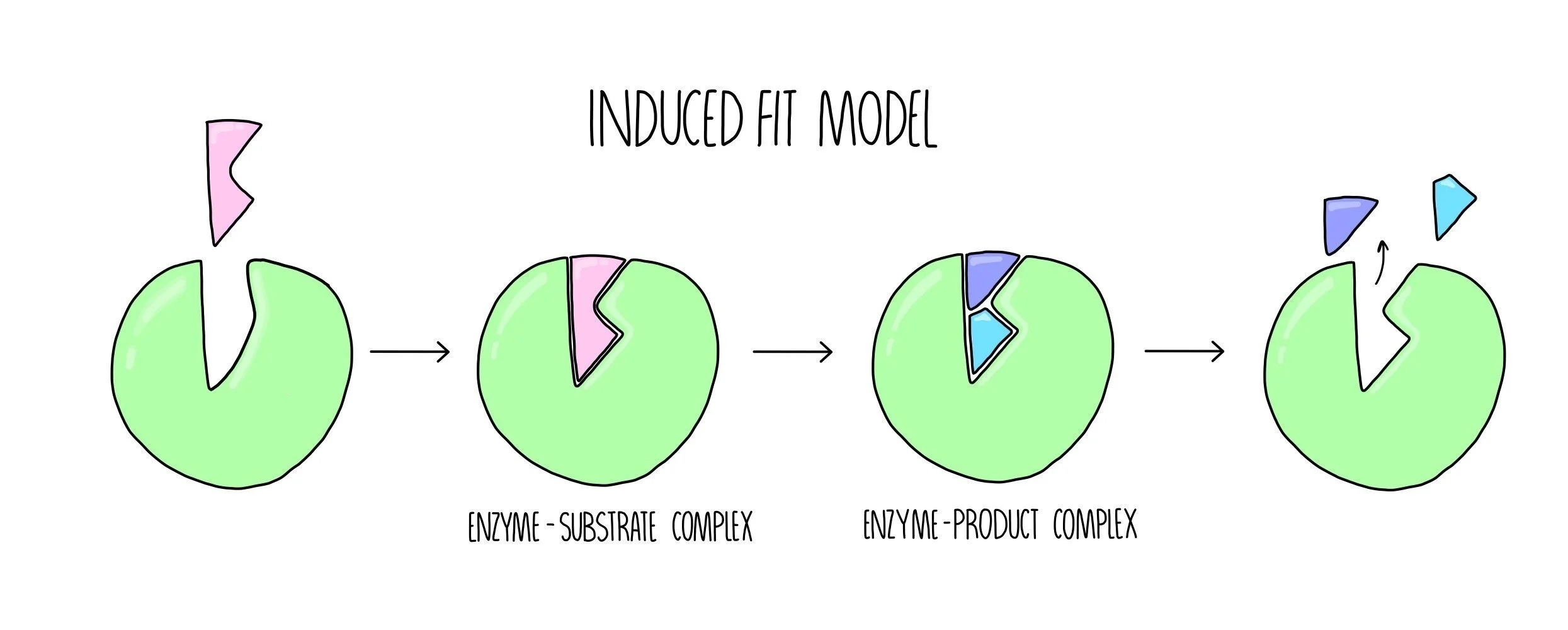
The induced fit model
The induced fit model suggests that the shapes of the enzyme’s active site and its substrate are not exactly complementary, but when the substrate enters the active site, a conformational change (change of shape) occurs which induces catalysis. The induced fit model can be broken down into the following stages:
The substrate enters the enzyme’s active site, forming an ES complex.
The enzyme undergoes a conformational change which causes the conversion of substrate into product, forming an EP complex.
The product is released from the enzymes active site.
Comparing the two models of enzyme action
The advantage of the lock-and-key model is that it explains why most enzymes display such high specificity to their substrates. Each enzyme will catalyse only a certain type of reaction and will only bind to a single specific substrate out of the millions of different molecules that are floating around our bodies. However, not all enzymes catalyse a single chemical reaction. For example, lipase exhibits broader specificity and can bind to a variety of lipids, which only the induced fit model is able to explain. In addition, the induced fit model is better able to explain how catalysis actually occurs. A conformational change, which would place stress on the bonds within the substrate can explain how bonds would break in order for the products to form. This makes the induced fit model the more widely accepted model of the two.
Factors which affect rate of reaction
Enzyme concentration
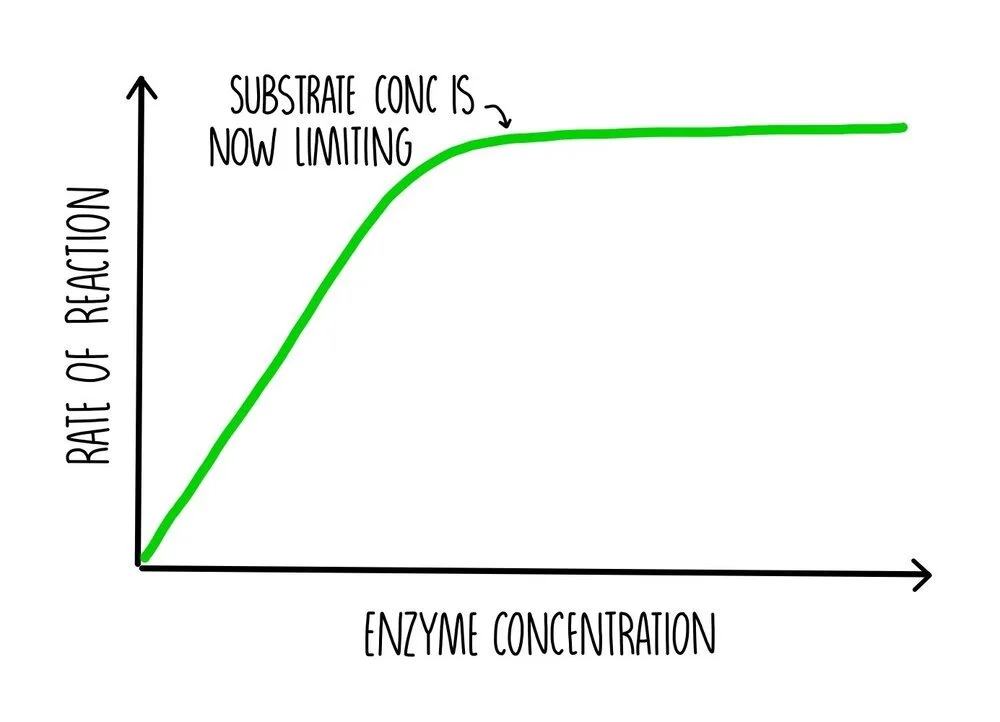
As enzyme concentration increases, the rate of reaction increases since more active sites will be available to bind to substrate molecules. This means that there will be more frequent collisions between the enzyme and substrate, so there will be more formation of enzyme-substrate complexes. However, a point will be reached when increasing enzyme concentration does not result in further increases in reaction rate. At this point, something else has become a limiting factor, such as the availability of substrate.
Substrate concentration
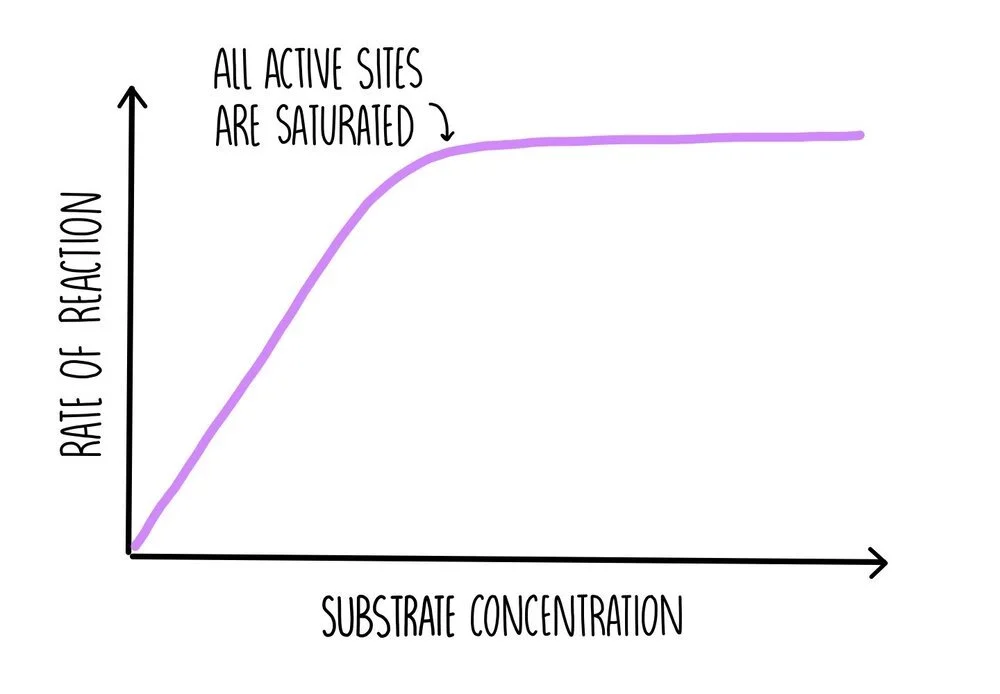
As substrate concentration increases, the rate of reaction increases since there are more substrate molecules to fill the enzyme’s active sites. There will be more frequent collisions so more formation of ES complexes. At some point a ‘saturation’ point is reached where all of the enzyme’s active sites are occupied with substrate molecules, so the addition of more substrate molecules will have no effect on the rate of reaction. At this point, the reaction is proceeding as fast as possible, which is referred to as Vmax. The only way the reaction can go any faster is by increasing enzyme concentration.
Temperature
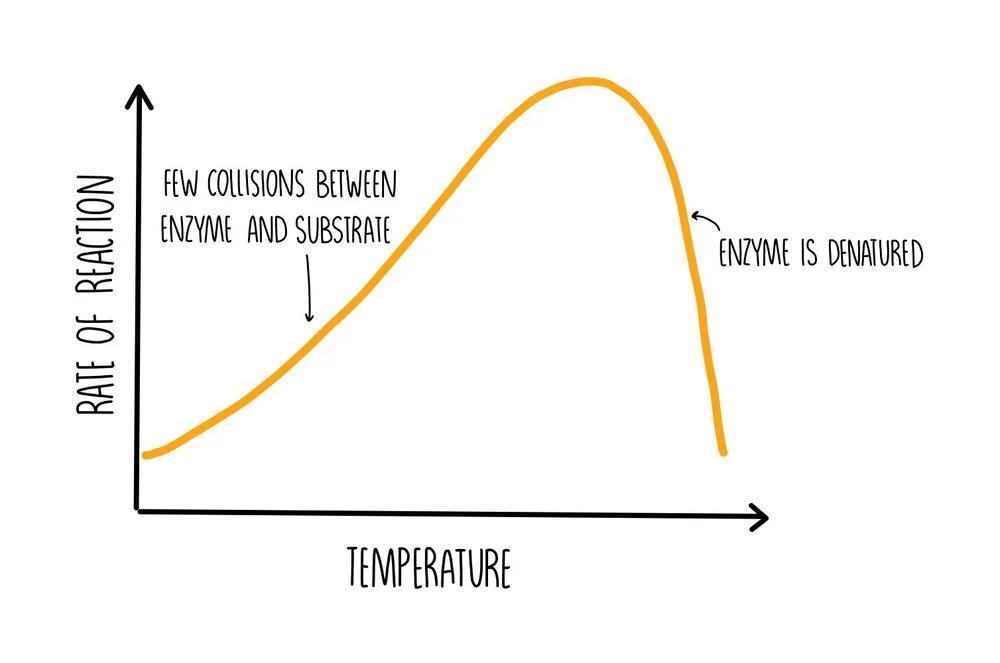
At low temperatures, the rate of reaction will be slow because the enzyme and substrate have low amounts of kinetic energy. This means that there won’t be many collisions so there will be reduced formation of ES complexes. As the temperature is increased, the number of collisions increases, increasing the formation of ES complexes and increasing the rate of reaction. If the temperature becomes really high, hydrogen bonds will begin to break within the protein, causing it to unravel and become denatured. If enzymes are denatured, they lose the shape of their active sites which means they cannot bind to their substrate, decreasing the rate of reaction.
pH
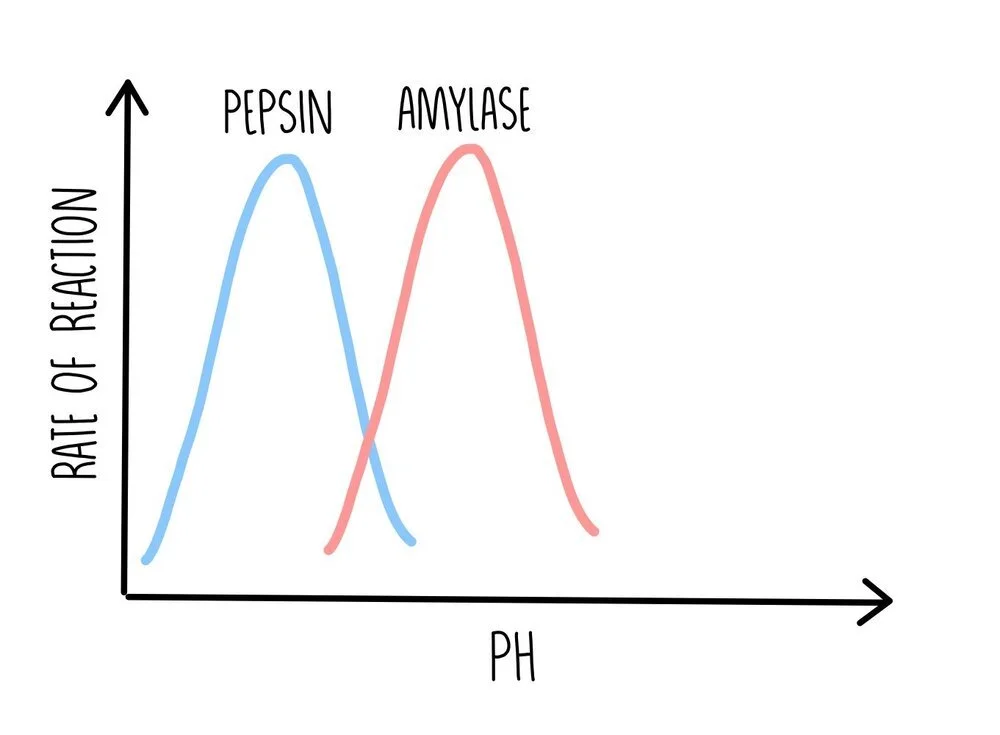
Each enzyme has its own optimum pH at which it works best. Pepsin, the enzyme which digests protein in the stomach, works best in acidic environments whereas the enzymes responsible for the digestion of carbohydrates work better at a more neutral pH. Deviations from the optimum pH change the charge on the enzyme, which affects ionic bonding within its structure. Deviations in pH also break hydrogen bonds. This causes it to change shape and become denatured, decreasing the rate of reaction as pH deviates from the enzyme’s optimum conditions.
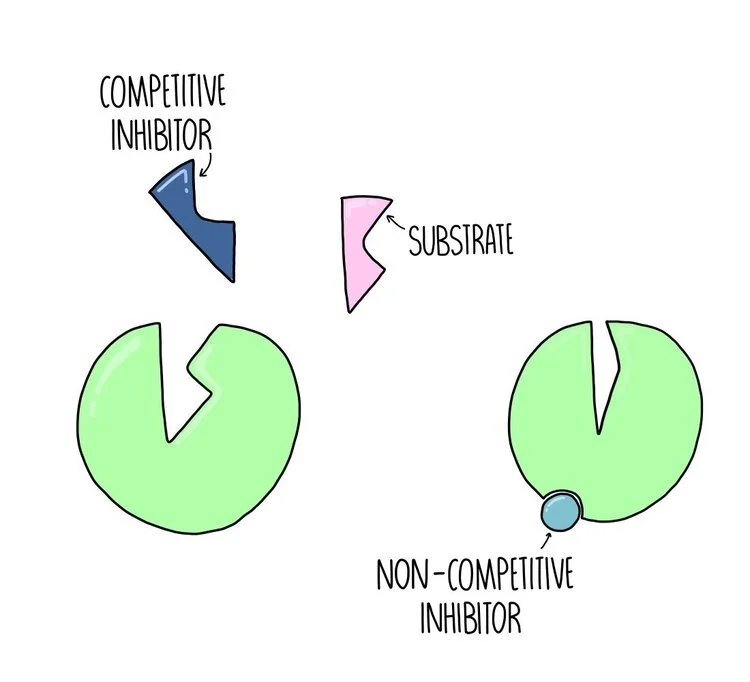
Competitive vs non-competitive inhibitors
Competitive inhibitors are ones which bind to the active site of the enzyme, blocking the substrate from binding - they compete with the substrate for access to the enzyme’s active site. The effect of a competitive inhibitor can be reduced by increasing substrate concentration. If there is way more substrate compared to inhibitor then the substrate is much more likely to collide with the enzyme’s active site.
Non-competitive inhibitors are ones which bind to a site on the enzyme away from its active site. This region is known as its allosteric site. The effect of a non-competitive inhibitor cannot be reduced by increasing substrate concentration.
Investigating the rate of an enzyme-controlled reaction
There are two ways of measuring the rate of an enzyme controlled reaction: By measuring the speed at which product is formed or reactant is removed.
Measuring product formation using catalase
A good example of a reaction in which product formation is measured is the reaction catalysed by catalase, which breaks down hydrogen peroxide into oxygen and water.
Method:
Add a set volume/concentration of hydrogen peroxide to a test tube along with a set volume of buffer solution to control pH.
Place a bung on top of the test tube and connect to an inverted measuring cylinder using delivery tubing.
Using a pipette, add catalase the test tube. Quickly re-insert the bung and delivery tube.
Measure the volume of oxygen produced in the measuring cylinder every ten seconds for the first minute of the reaction (using a stopwatch to record the time).
Repeat two more times. Calculate the mean volume of oxygen produced at each ten second time interval.
Control variables: volume of catalase, volume and concentration of hydrogen peroxide solution, pH, temperature.
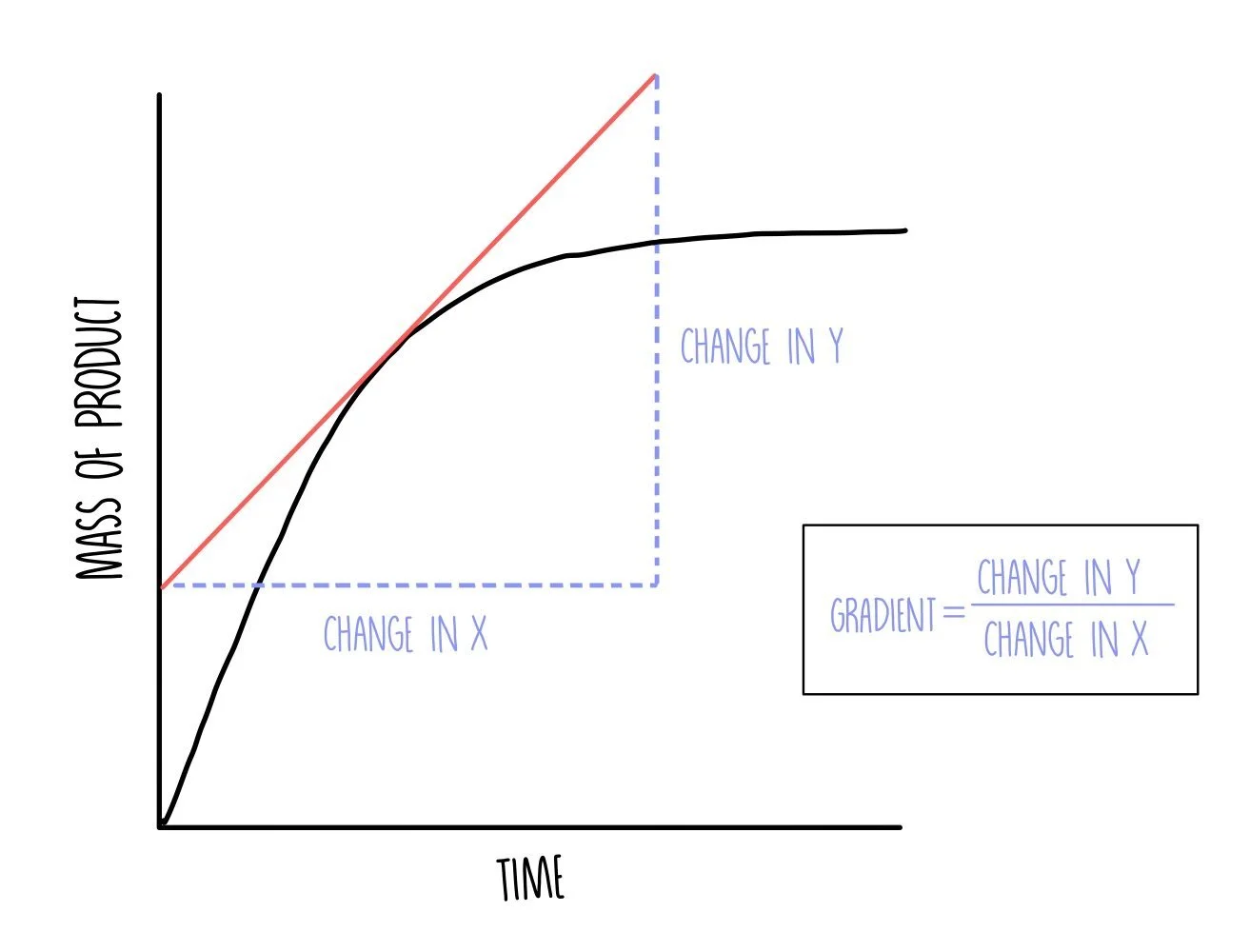
Plot a graph with time on the x-axis and volume of oxygen on the y-axis. You can determine the initial reaction rate by drawing a tangent to the curve at t=0. Draw a straight line against the curve right at the beginning of the reaction and calculate the gradient by dividing the change in y by the change in x.
You can repeat the experiment with different concentrations of catalase to determine the effect of changing enzyme concentration on the initial rate of reaction.
Measuring reactant removal using amylase
Amylase is the enzyme which hydrolyses starch into maltose. You can measure decreasing starch concentration as the reaction progresses by adding iodine, which turns blue/black in the presence of starch.
Method:
Place a cuvette containing iodine solution in a colorimeter with a red filter. Zero the machine.
In a separate cuvette, pipette a given volume (e.g. 0.5 ml) of starch solution and a set volume of iodine solution. Mix together – the solution should turn blue/black. Place in the colorimeter and record the absorbance.
Add amylase solution of a set concentration/volume into the same cuvette and starch the stopwatch.
Record the absorbance of the solution every ten seconds.
Repeat the whole process two more times and calculate a mean absorbance for each ten second interval.
Control variables: concentration of iodine solution, volume of starch, pH, temperature.
Plot a graph with time on the x-axis and absorbance on the y-axis. You can repeat the experiment with difference concentrations of starch to determine the effect of changing substrate concentration on initial reaction rate.